NMDP [002 SP_B]
Vidhi Lalchand, Ph.D.
IMU Biosciences
13th Feb, 2026
[203 donors]
Data Semantics & Processing
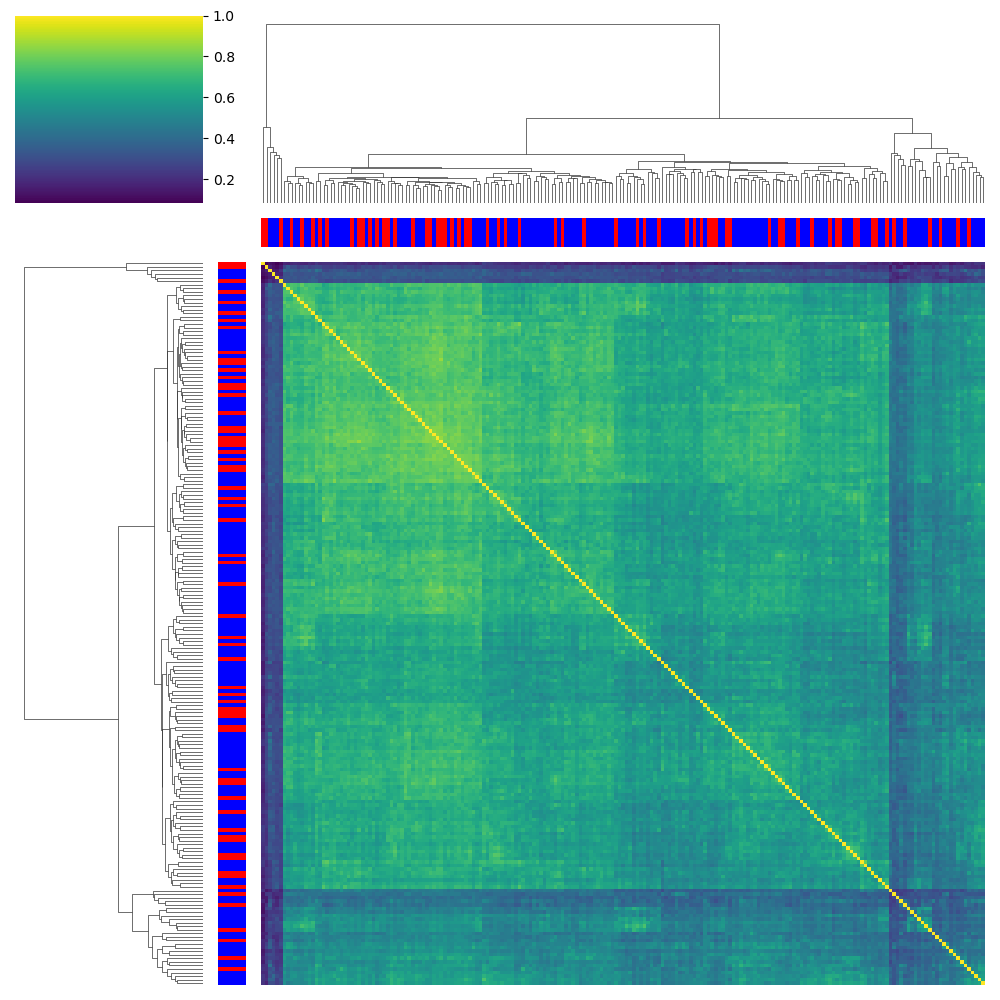
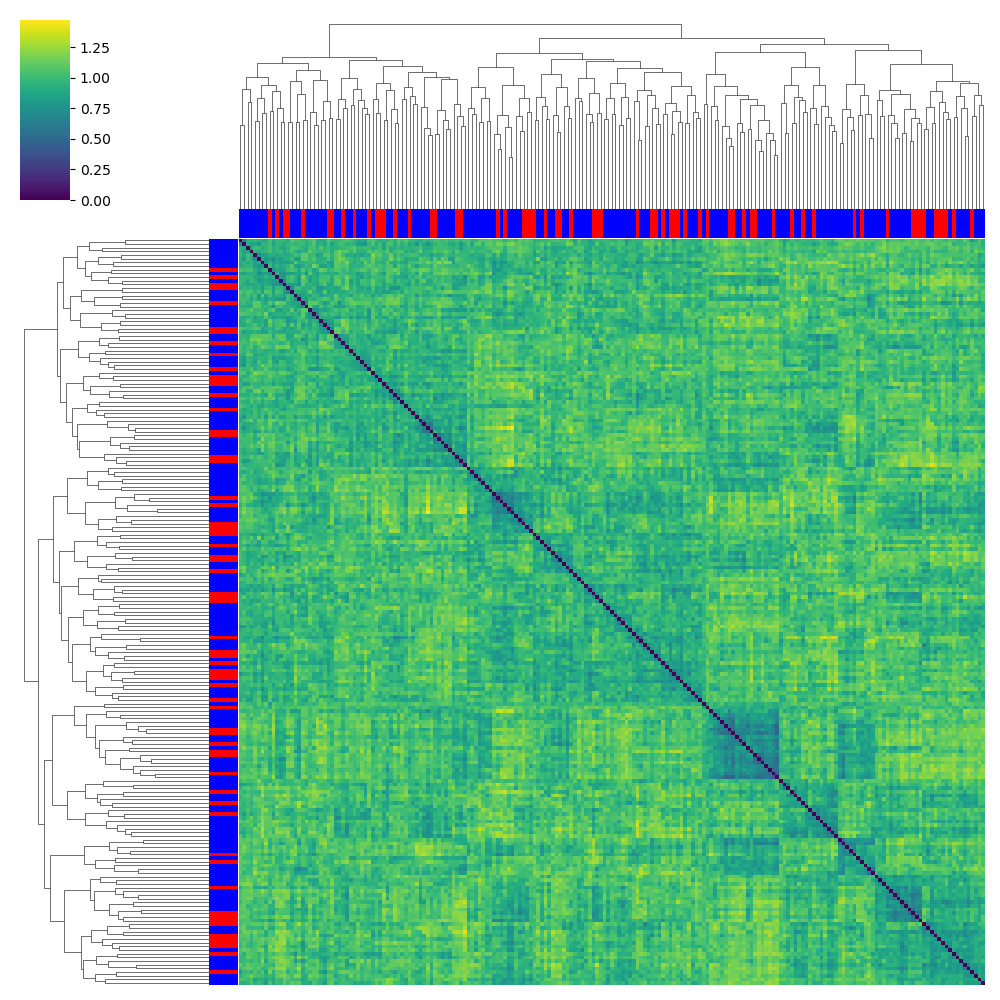
NMDP 002 SP_B Combined Ratios: Tv5 Panel
Columns dropped due to NaNs for significant cross-section of donors:
clean_tv_num = clean_tv_num.loc[:, clean_tv_num.nunique(dropna=False) > 1] # Drop columns with only one unique value
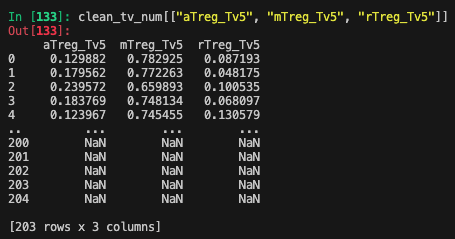
clean_tv_num = clean_tv_num.drop(columns=["aTreg_Tv5", "mTreg_Tv5", "rTreg_Tv5"])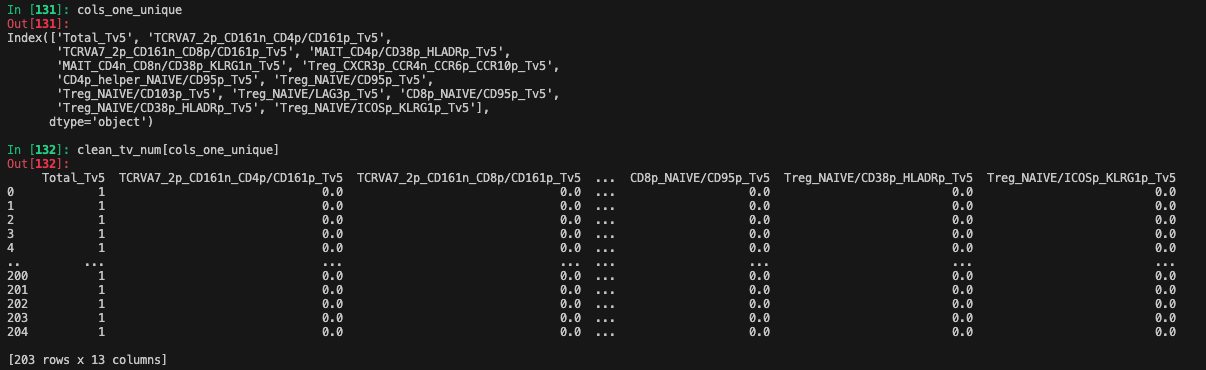
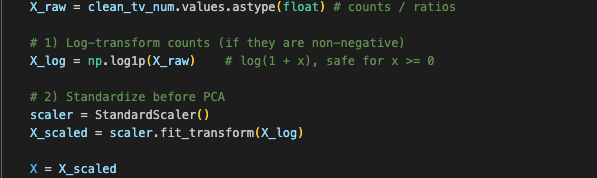
Log1p + Standardization
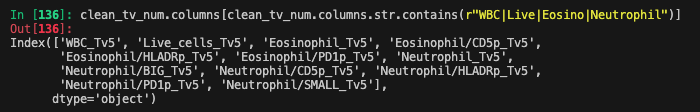
Batch effect + Noise columns
Off-the-shelf algorithms: 10-CV sweep
| Algorithm | % Acc | AUC | % Pos. class |
|---|---|---|---|
| Kernel Logistic Reg. | 0.5821 +/- 0.133 | ~0.5224 | 0.451 +/ 0.2 |
| Logistic Reg. | 0.5664 +/- 0.214 | ~0.5219 | 0.343 +/- 0.2 |
| RF Estimator | 0.665 +/- 0.077 | ~0.517 | < 0.20 |
| MLP + Dropout | 0.579 +/- 0.053 | ~0.58 | 0.455 +/ 0.2 |
| Boosting + XTrees | 0.670 +/- 0.042 | ~0.530 | < 0.20 |
| SVC | 0.606 +/- 0.167 | 0.511 | < 0.20 |
| GradientBoosting | 0.626 +/- 0.099 | ~0.49 | < 0.20 |
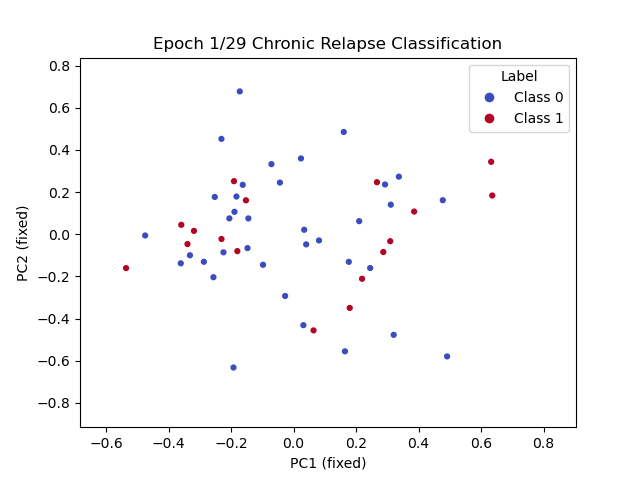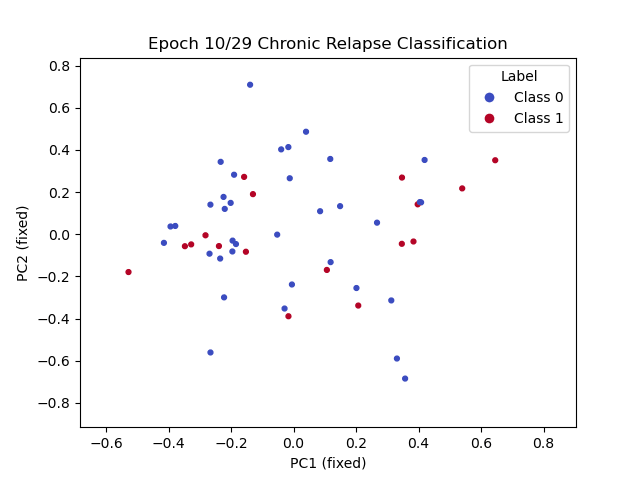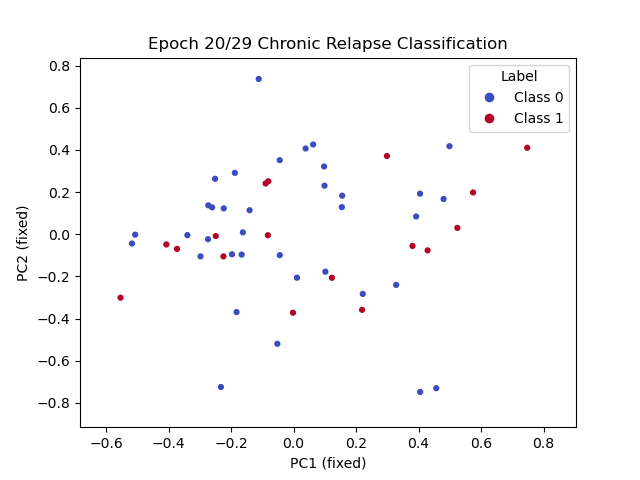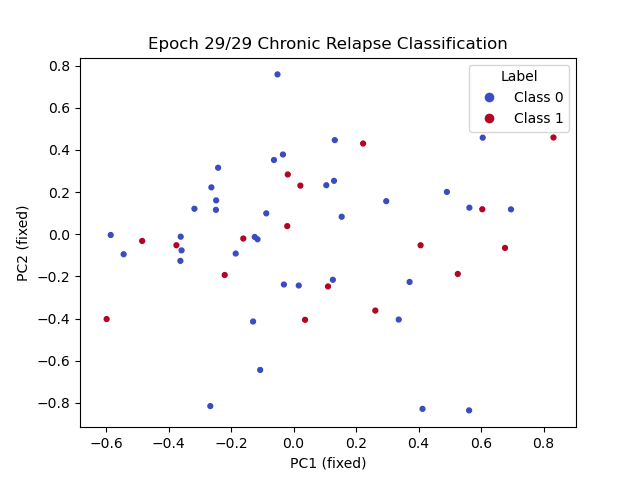
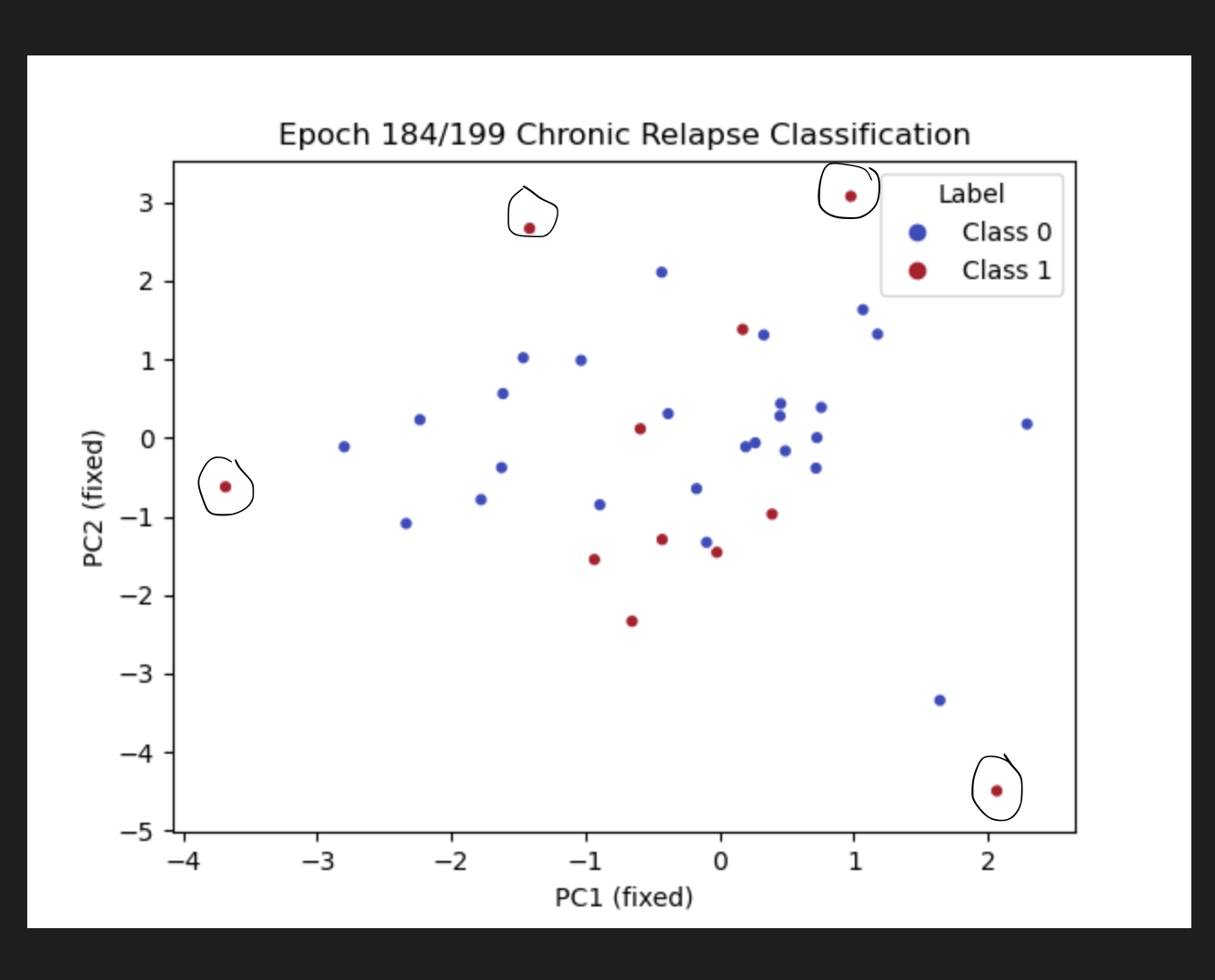
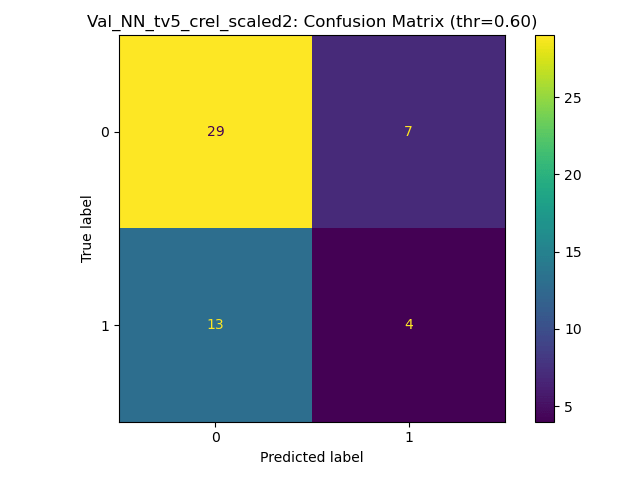
Goal: Predicting the binary response variable of chronic relapse {0,1} from the megatables (ratios). Unseen metrics with std. error of the mean (metric) for stable config:
Key failure mode: The fraction of chronic relapse donors correctly picked up by the model.
The AUC and % Accuracy obscure positive recall - which is the key thing to look out for here.
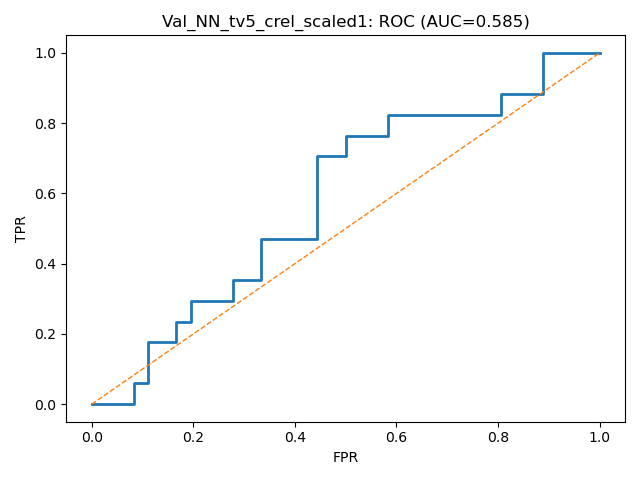
MLP + Dropout / w. imbalanced class weighting
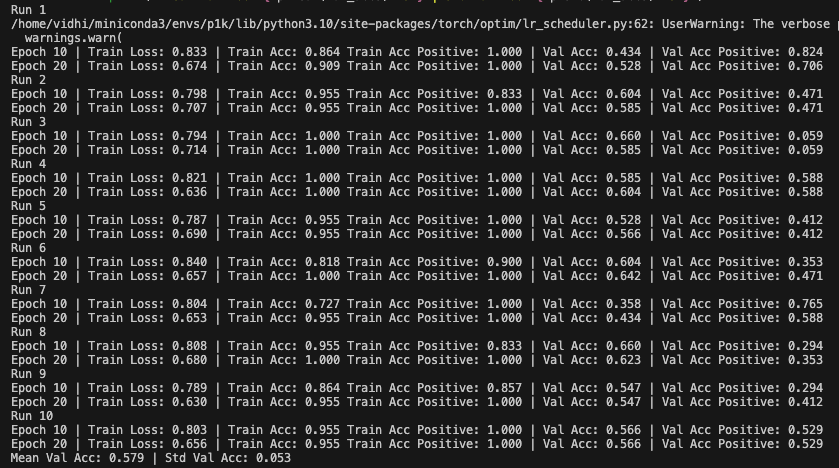
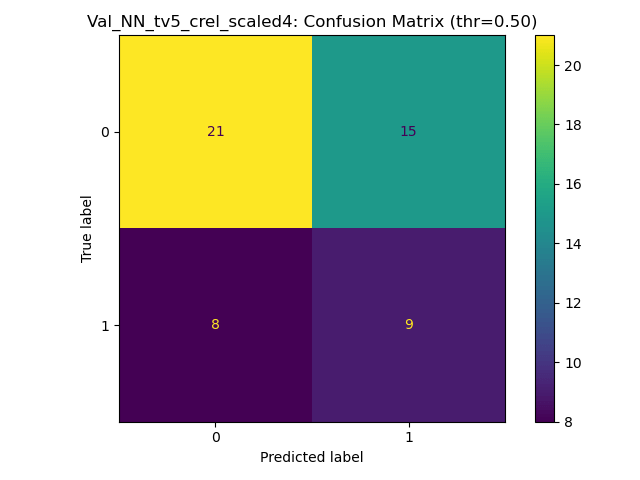
SP_A - Validation donors
SP_B - Validation donors

Reserved if time